
OmegaFold vs AlphaFold: Which AI is Better for Protein Structure Prediction?
Explore the detailed comparison of OmegaFold vs AlphaFold—two groundbreaking AI models in protein structure prediction. Learn about their architectures, accuracy, speed, technical strengths, and ideal use cases in modern biology and drug discovery.
HEALTH/DISEASEAI/FUTUREEDITOR/TOOLS
Sachin K Chaurasiya
4/13/20254 min read


In the ever-evolving field of computational biology, protein structure prediction stands as one of the most complex yet critical challenges. The emergence of AlphaFold by DeepMind and OmegaFold by Helixon marks a transformative moment in how we understand the architecture of proteins. These two AI models have set a new benchmark, offering researchers the ability to predict 3D protein structures with astonishing accuracy. But how do these two giants compare? In this article, we’ll explore OmegaFold vs. AlphaFold—their foundations, performance, strengths, limitations, and what makes each of them unique in decoding life’s molecular puzzles.
What is AlphaFold?
AlphaFold, developed by DeepMind, is an AI-based system that predicts a protein's 3D structure from its amino acid sequence. Released in 2020 and later enhanced with AlphaFold2, it created a seismic shift in structural biology by achieving accuracy comparable to experimental methods like X-ray crystallography and cryo-EM. The AlphaFold Protein Structure Database now hosts over 200 million protein structures, revolutionizing access to structural information.
What is OmegaFold?
OmegaFold, launched by Helixon in 2022, offers a novel approach to structure prediction. Unlike AlphaFold, it does not rely on Multiple Sequence Alignments (MSAs), which are time-consuming and resource-heavy. Instead, OmegaFold uses a language model trained on protein sequences, delivering high-quality predictions with remarkable speed and efficiency. It targets a broader spectrum of proteins, especially those with limited evolutionary data.
Architectural Comparison: AlphaFold vs OmegaFold
AlphaFold: An End-to-End Deep Learning Pipeline
AlphaFold2 is a complex architecture combining deep learning and structural bioinformatics. Its key component is the Evoformer, a novel transformer-based module that operates on two tracks: a Multiple Sequence Alignments (MSA) track and a pairwise residue track. The Evoformer performs iterative reasoning using triangle multiplicative updates, attention mechanisms, and outer product transformations to capture both local and long-range dependencies.
AlphaFold utilizes templates from the Protein Data Bank (PDB) and MSA data as inputs, feeding them through these advanced networks to produce a spatial graph representing the protein backbone. The network is trained with a loss function that includes
Frame-aligned point error (FAPE)
Violation loss for physical plausibility
Predicted Local Distance Difference Test (pLDDT)
Predicted aligned error (PAE)
OmegaFold: MSA-Free Transformer with Structure Decoder
OmegaFold takes a minimalist yet highly effective approach. It discards MSAs in favor of a large protein language model (pLM) trained on UniRef50 and BFD datasets, learning from evolutionary patterns embedded in sequence data alone. It employs rotary position embeddings, axial attention, and residual blocks to encode contextual dependencies within the sequence.
The architecture includes
A pretrained transformer-based protein encoder (akin to ESM or ProtBert)
A geometry decoder module that predicts torsion angles, distances, and pairwise atom coordinates
An inverse kinematics-based reconstruction step to derive full-atom 3D structures
OmegaFold uses end-to-end differentiable modeling to minimize deviations in predicted coordinates from true PDB structures, relying on coordinate-based loss functions rather than alignment-based ones.


Speed & Efficiency
AlphaFold can take hours to predict a structure due to its reliance on large-scale MSA searches, template lookups, and large model inference. It typically requires multi-GPU setups or high-memory cloud instances.
OmegaFold bypasses MSAs entirely, dramatically reducing computational time. On average, a single structure prediction can be completed in under 10 minutes using a single A100 GPU, making it up to 20x faster than AlphaFold in practice.
Accuracy and Performance Metrics
AlphaFold Accuracy
Average GDT_TS score: >90 for CASP14 targets
pLDDT > 90 in 58% of predicted proteins
Exceptional at modeling globular domains and proteins with rich evolutionary data
Limitations in modeling intrinsically disordered proteins and large multi-domain assemblies
OmegaFold Accuracy
GDT_TS ~85–90 for benchmark targets
Outperforms AlphaFold in orphan proteins (no known homologs)
Comparable performance for single-domain and short proteins
Slight drop in accuracy for complex, multi-chain systems
Benchmark Evaluations
On CAMEO datasets, AlphaFold performs better on well-studied proteins with evolutionary data, but OmegaFold delivers more reliable results for novel or synthetic proteins.
RMSD deviations between OmegaFold and experimental structures are ~2.0–2.5 Å, while AlphaFold often achieves <1.5 Å RMSD.
Application Areas
AlphaFold
Structural genomics
Antibody-antigen interaction modeling
Enzyme-substrate complex prediction
Rational drug design via virtual screening
OmegaFold
High-throughput protein screening
Rapid modeling of metagenomic data
Synthetic biology and protein engineering
Initial structure modeling for downstream AlphaFold refinement
Experimental Validation and Real-World Use
AlphaFold structures have been experimentally validated using
Cryo-EM overlays
NMR-derived constraints
Crystallographic RMSD comparisons
Molecular dynamics (MD) stability simulations
OmegaFold predictions are being actively validated with
AlphaFold overlay comparisons
Ligand-binding pocket prediction accuracy
Folding energy landscapes using Rosetta Relax
Integration with RNA-binding and post-translational modification prediction tools
Community and Ecosystem
AlphaFold
Backed by DeepMind and EMBL-EBI
AlphaFold Database with 200M+ structures
Extensions like AlphaFold-Multimer and ColabFold
Integrated into ChimeraX, PyMOL, and Rosetta
OmegaFold
GitHub project maintained by Helixon
Lightweight Docker images for deployment
API endpoints for integration into proteomics pipelines
Gaining adoption in ML-first biotech companies
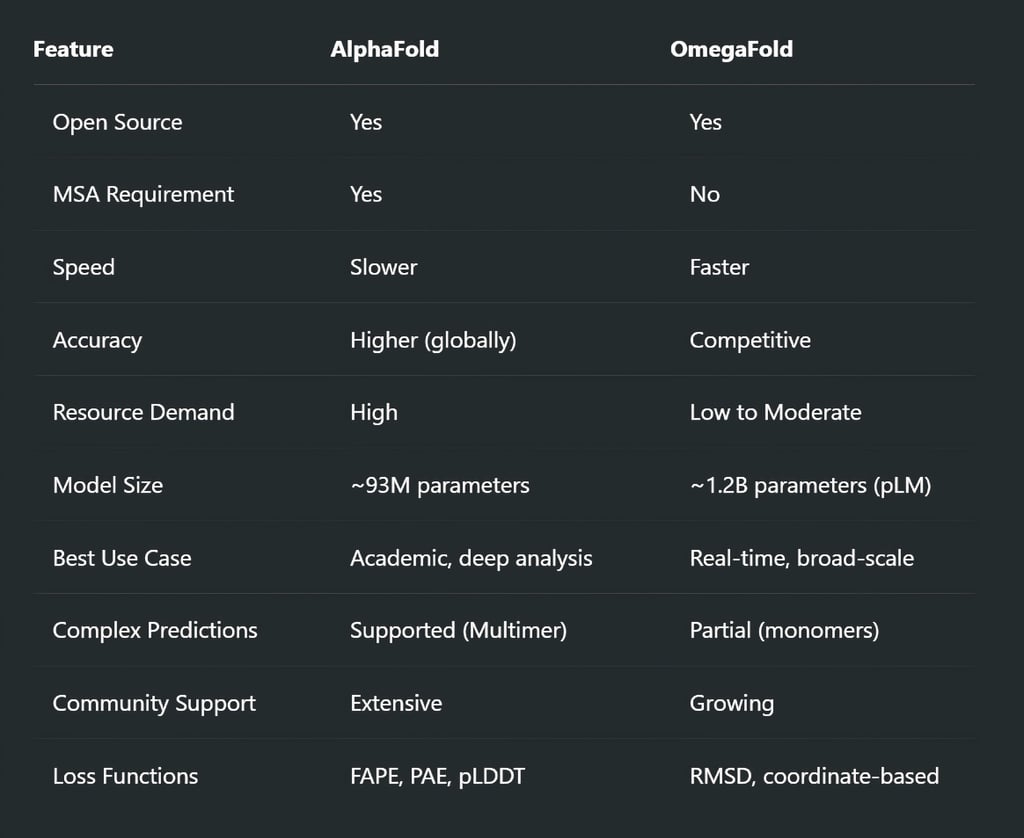
The Verdict: Which One Should You Use?
It’s not a matter of AlphaFold vs OmegaFold—it’s about the context of use.
Use AlphaFold when:
You need top-tier accuracy and structural completeness
Modeling large or complex protein assemblies
You have access to high-performance GPUs and storage
Use OmegaFold when:
You need fast, good-enough models
Working with novel, synthetic, or MSA-poor sequences
You're deploying at scale or working with limited hardware
AlphaFold has indisputably transformed protein structure prediction with unmatched accuracy and a comprehensive database. OmegaFold, however, brings accessibility and speed to the forefront, enabling broader adoption across scientific domains. Together, they form a powerful duo, driving the future of molecular biology and accelerating discoveries in health, disease, and beyond.
As AI-driven science continues to advance, tools like AlphaFold and OmegaFold remind us of the extraordinary potential of blending biological insight with machine intelligence. Whether you're a biologist, data scientist, or innovator, the protein structure revolution is here—and it’s just getting started.
Subscribe To Our Newsletter
All © Copyright reserved by Accessible-Learning Hub
| Terms & Conditions
Knowledge is power. Learn with Us. 📚


