
OmegaFold vs AlphaFold: Revolutionizing Protein Structure Prediction?
Explore the in-depth comparison between OmegaFold and AlphaFold, two groundbreaking AI models transforming protein structure prediction. Learn about their unique features, applications, limitations, and how they complement each other in advancing biological research, drug discovery, and synthetic biology.
AI/FUTUREHEALTH/DISEASECOMPANY/INDUSTRYEDUCATION/KNOWLEDGE
Sachin K Chaurasiya
1/18/20256 min read
The scientific community has long recognized the immense importance of understanding protein structures. Proteins are the workhorses of biology, playing critical roles in virtually every cellular process. Predicting their structures is key to unlocking advancements in medicine, drug development, and synthetic biology. Among the most innovative solutions to this challenge are AlphaFold and OmegaFold—two groundbreaking AI systems that have revolutionized our understanding of protein folding. In this article, we’ll explore the nuances of these models, their capabilities, and how they differ from one another.
The Rise of Protein Structure Prediction
Before diving into the specifics of Alphafold and OmegaFold, it’s crucial to understand the broader problem. Proteins are composed of amino acid chains that fold into unique three-dimensional structures, dictating their function. Predicting these structures from a sequence of amino acids (known as the protein-folding problem) has been a central challenge in biology for decades.
Traditional methods, like X-ray crystallography and cryo-electron microscopy, though accurate, are resource-intensive and time-consuming. Computational approaches, including AI-based systems, have emerged as game changers, providing faster and often equally accurate predictions.
AlphaFold: The AI Powerhouse from DeepMind
AlphaFold, developed by DeepMind (a subsidiary of Alphabet), is a machine learning model that stunned the scientific world with its exceptional performance in the CASP (Critical Assessment of protein Structure Prediction) competition. It uses a combination of deep learning techniques and biological knowledge to predict protein structures with remarkable accuracy.
Key Features
Transformer Architecture: AlphaFold employs a deep neural network to interpret multiple sequence alignments (MSAs) and predict protein structures.
High Accuracy: In CASP14 (2020), AlphaFold achieved an average global distance test (GDT) score of over 90, rivaling experimental methods.
Versatility: AlphaFold has been particularly successful in predicting single-chain protein structures, making it a robust tool for fundamental research.
Open Access: DeepMind released AlphaFold’s code and predictions for the human proteome, democratizing access to high-quality protein structure data.
Human Proteome Coverage: AlphaFold has predicted structures for nearly the entire human proteome, aiding research in personalized medicine and rare diseases.
Limitations
AlphaFold has been hailed as a revolutionary tool for drug discovery, enzyme design, and understanding diseases caused by misfolded proteins, like Alzheimer’s.
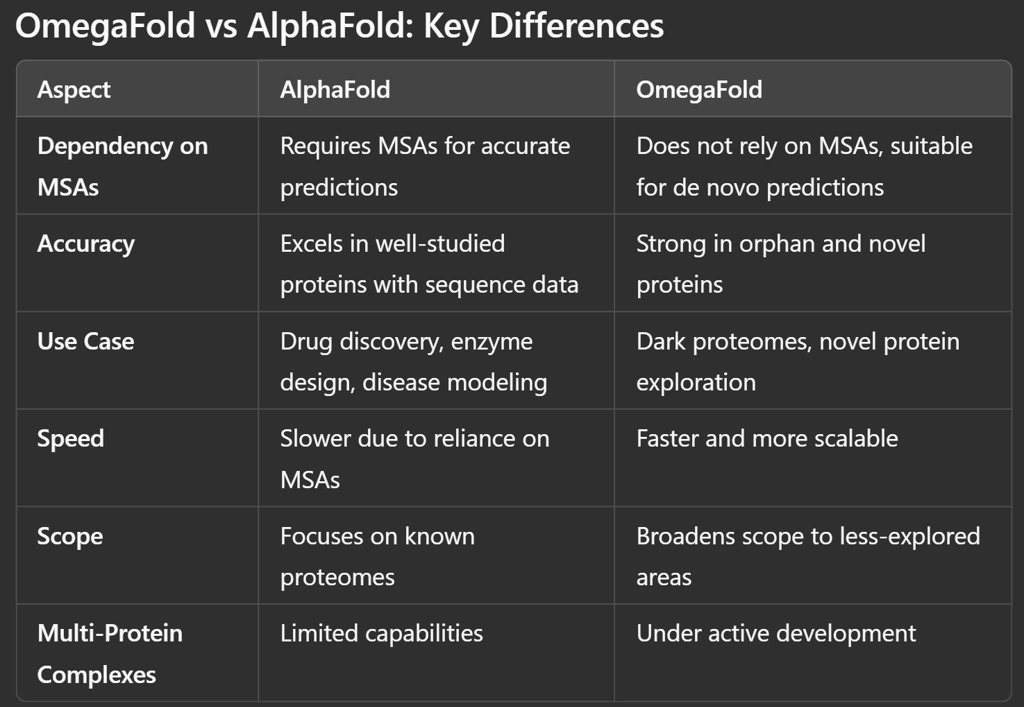
Dependence on MSAs: AlphaFold relies heavily on evolutionary data, limiting its effectiveness for proteins without known homologs.
Multi-Protein Complexes: While AlphaFold can predict single proteins accurately, predicting multi-protein interactions remains a challenge.
Dynamic Proteins: It struggles to capture the flexibility and dynamics of proteins in different environments or states.
Applications in the Real World
Drug Discovery: AlphaFold has been instrumental in identifying drug targets for diseases like malaria and cancer.
Biological Research: By decoding structures of nearly the entire human proteome, AlphaFold has set the stage for countless scientific breakthroughs.
Disease Modeling: Its ability to predict structures of misfolded proteins has profound implications for neurodegenerative diseases.
Evolutionary Insights: AlphaFold’s reliance on MSAs has provided insights into evolutionary relationships between proteins.


OmegaFold: A Leap into De Novo Predictions
While AlphaFold’s achievements are undeniable, its reliance on MSAs and evolutionary data limits its applicability for some protein classes, particularly those with limited sequence homology. Enter OmegaFold, a deep learning model by HelixAI designed to tackle these limitations head-on.
Key Features
No Dependency on MSAs: Unlike AlphaFold, OmegaFold does not require evolutionary information or alignments, making it suitable for de novo predictions.
Speed and Scalability: OmegaFold offers faster predictions, crucial for large-scale applications and real-time analyses.
Focus on Orphan Proteins: OmegaFold excels in predicting structures for proteins with no known homologs, expanding the scope of structural biology into uncharted territory.
Broader Utility in Genomics: Its ability to predict novel proteins opens doors to exploring dark proteomes—areas of the genome encoding poorly understood proteins.
Adaptive Learning: OmegaFold leverages advanced deep learning techniques to handle diverse protein sequences, making it highly adaptable.
Limitations
Lower Accuracy for Well-Studied Proteins: For proteins with extensive evolutionary data, OmegaFold’s predictions may not be as precise as AlphaFold’s.
Limited Benchmarking: OmegaFold’s performance has not been as extensively validated as AlphaFold’s in competitive settings like CASP.
Complex Systems: Its ability to predict protein-protein interactions and larger complexes is still under development.
Applications in the Real World
Uncharted Proteins: OmegaFold’s ability to predict structures without MSAs has opened the door to studying obscure proteins and pathogens.
Synthetic Biology: It aids in designing novel proteins for industrial applications, such as biofuels and biodegradable plastics.
Exploration of Dark Proteomes: OmegaFold’s focus on orphan proteins allows researchers to delve into previously unexplored regions of genomes, potentially uncovering new biological functions.
Precision Agriculture: OmegaFold’s insights into plant proteomes can aid in developing resilient crops and sustainable agricultural practices
Accelerating the Field of Proteomics
Proteomics, the study of all proteins within an organism, has historically been constrained by the speed of experimental structure determination methods. AI models like AlphaFold and OmegaFold significantly reduce this bottleneck, making proteome-scale studies possible in weeks instead of years. This has profound implications for:
Biomedical research: Targeting disease-specific proteins for therapy.
Epidemiology: Understanding the structural aspects of viral proteomes, especially during outbreaks like COVID-19.
Systems biology: Mapping protein networks within cells for deeper insights into cellular functions.
AI for Understanding Protein Engineering & Design
OmegaFold and AlphaFold are catalyzing the field of protein engineering by enabling scientists to predict how modifications to protein sequences will affect their structure and function. This facilitates:
Custom enzyme design: For industrial applications like biofuel production and waste recycling.
Protein therapeutics: Creating proteins with enhanced stability or specificity for medical applications.
Synthetic biology: Designing entirely new proteins for novel functions, such as biosensors.
Beyond Proteins: Applications to RNA and Complex Molecules
While AlphaFold focuses on proteins, researchers are adapting its architecture to predict RNA folding and interactions between proteins and other biomolecules. RNA structure prediction is particularly important for:
Targeting RNA viruses (e.g., SARS-CoV-2): Understanding RNA folding can aid in antiviral drug design.
RNA-based therapeutics: Such as small interfering RNA (siRNA) or mRNA-based vaccines.
RNA-protein complexes: Studying complexes like the ribosome to reveal how genetic information is translated into proteins.
Democratization of Structural Biology
Tools like AlphaFold and OmegaFold lower barriers for entry into structural biology research. Labs that previously couldn’t afford expensive techniques like X-ray crystallography or cryo-EM now have access to high-quality structure predictions with minimal computational resources. This democratization:
Levels the playing field for resource-limited regions.
Spurs innovation in countries without significant investment in high-tech research infrastructure.
Enables interdisciplinary research, as non-biologists (e.g., data scientists or physicists) can now contribute to protein-related discoveries.
Ethical and Societal Considerations
Dual-use concerns: Misuse of protein prediction tools for designing harmful biological agents.
Equity in access: Ensuring open access to such tools so developing nations and underfunded labs can benefit.
Job displacement: As AI takes on labor-intensive prediction tasks, what roles will traditional experimental structural biologists play?
Reliability: Over-reliance on AI models may lead to assumptions that aren’t fully validated, especially for systems where predictions are less accurate.
The Role of Benchmarking & Independent Validation
Although both AlphaFold and OmegaFold are highly promising, their predictions still require benchmarking against experimental results. This is particularly important for:
Dynamic proteins: Many proteins adopt multiple conformations, and AI often predicts only the most stable structure.
Non-protein components: AI struggles to predict interactions with ions, small molecules, or membranes, which are critical in many biological contexts.
Challenges and Future Directions
While AlphaFold and OmegaFold are milestones in AI-driven biology, challenges remain:
Dynamic Structures: Proteins are not static; understanding their dynamics, conformational changes, and interactions is the next frontier.
Complex Systems: Predicting multi-protein complexes and interactions with non-protein molecules (e.g., DNA, RNA, small molecules) remains a significant challenge.
Data Integration: Combining AI predictions with experimental data (e.g., cryo-EM maps) can enhance reliability and accuracy.
Scalability: As the demand for large-scale protein predictions grows, optimizing computational efficiency will be crucial.
Broader Accessibility: Ensuring that both models are accessible to researchers in resource-limited settings is essential for global scientific progress.


AlphaFold and OmegaFold represent two sides of the same coin: one excels in leveraging evolutionary data for well-studied proteins, while the other shines in de novo predictions, pushing the boundaries of structural biology. Together, they embody the transformative power of AI in science, enabling researchers to explore the intricate world of proteins faster and more accurately than ever before.
As these tools continue to develop, they will undoubtedly drive new discoveries, advancing our understanding of life and reshaping fields like medicine, agriculture, and biotechnology. The future of protein science has never looked brighter, thanks to these AI pioneers.
Subscribe To Our Newsletter
All © Copyright reserved by Accessible-Learning Hub
| Terms & Conditions
Knowledge is power. Learn with Us. 📚


