
ESMFold vs AlphaFold Explained: Speed, Accuracy, and Real-World Impact
Explore the in-depth comparison between ESMFold and AlphaFold—two revolutionary AI models transforming protein structure prediction. Discover their architectures, use cases, accuracy, and how they’re reshaping biology and drug discovery.
MODERN DISEASESAI ASSISTANTCOMPANY/INDUSTRYHEALTH/DISEASEAI/FUTURE
Sachin K Chaurasiya
4/15/20253 min read

In recent years, the world of computational biology has witnessed groundbreaking innovations, especially in the field of protein structure prediction. Two major players making waves in this space are AlphaFold by DeepMind and ESMFold developed by Meta AI. Both aim to decode the complex structures of proteins from their amino acid sequences, but their methodologies and capabilities differ in compelling ways.
The Basics: Why Protein Structure Matters
Proteins are fundamental to biological functions. From catalyzing metabolic reactions to DNA replication, proteins are the unsung heroes of cellular machinery. Their functionality is determined not just by their amino acid sequence but by how this sequence folds into a 3D structure. Predicting this structure accurately has long been a challenge in biology—until AI stepped in.
Traditional experimental methods such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy are costly, time-consuming, and often limited by experimental constraints. AI-driven protein structure prediction offers a revolutionary alternative by reducing timelines from months to minutes, making it feasible to predict millions of protein structures at scale.
AlphaFold: The Game Changer
AlphaFold, developed by DeepMind, stunned the scientific community in 2020 when it demonstrated unmatched performance at CASP14 (Critical Assessment of protein Structure Prediction). What makes AlphaFold special is its use of deep learning to understand evolutionary patterns in protein sequences and map them to accurate 3D coordinates.
Key Features
Multiple Sequence Alignments (MSAs): AlphaFold leverages MSAs to derive evolutionary couplings that indicate residue-residue contacts.
Templates from solved structures: Integrates known protein templates from the PDB database for improved accuracy.
Attention-based Neural Network Architecture: The Evoformer module in AlphaFold enables complex reasoning over sequences and spatial relationships.
End-to-End Differentiable Model: Unlike traditional pipelines, AlphaFold is trained end-to-end, directly optimizing structural accuracy.
Open Database: The AlphaFold Protein Structure Database, in collaboration with EMBL-EBI, provides over 200 million protein structure predictions.
Technical Highlights
Utilizes inter-residue distance maps and torsion angle predictions for fine-grained modeling.
Employs recycling in the neural network, allowing it to iteratively refine predictions.
Capable of modeling disordered regions with confidence scores (pLDDT) indicating reliability.
AlphaFold's impact has been transformational in areas like drug discovery, evolutionary biology, and structural genomics.


ESMFold: Speed, Scalability, and Simplicity
In contrast, ESMFold (Evolutionary Scale Modeling Fold) by Meta AI is optimized for speed and scale. Its key innovation lies in eliminating the dependency on MSAs and structural templates. Instead, it relies on a transformer-based protein language model trained on massive datasets.
Key Features
No MSAs or templates required, dramatically reducing computational overhead.
Based on the ESM-2 (650M parameter) language model, which captures nuanced sequence relationships.
Generates 3D structures by coupling the transformer outputs with a structure module similar to AlphaFold's.
Integrated with Meta’s ESM Metagenomic Atlas, which features over 600 million protein predictions.
Technical Highlights
Uses masked language modeling (MLM) to pre-train on unlabeled protein sequences, capturing evolutionary signals inherently.
The transformer layers in ESMFold internally encode long-range dependencies, mimicking the effect of MSA-based coevolution.
Achieves a median TM-score > 0.7 for high-confidence predictions, competitive with AlphaFold.
Able to predict structures in seconds per sequence, enabling real-time applications.
ESMFold’s architecture positions it as a game-changer for researchers working with large, uncharacterized datasets, such as in environmental or microbiome research.
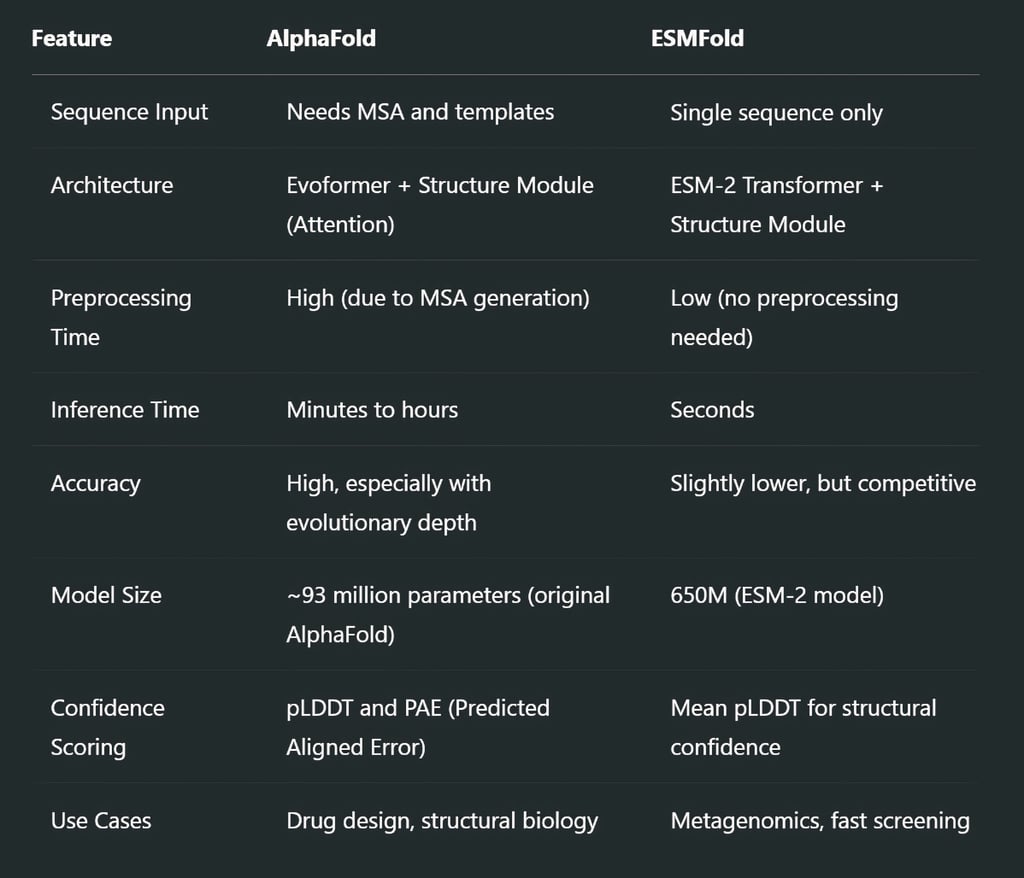
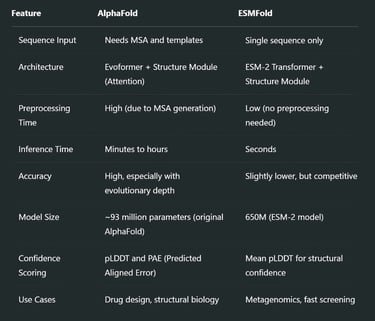
Complementary Strengths
Rather than seeing them as competitors, many researchers view AlphaFold and ESMFold as complementary tools. AlphaFold’s deep accuracy is invaluable for fine-grained structural insights, especially in clinical and pharmaceutical applications. ESMFold’s strength lies in its agility—ideal for fast, large-scale protein annotation and structure discovery.
A hybrid approach is gaining traction: using ESMFold for fast prototyping and AlphaFold for in-depth refinement. This workflow maximizes both speed and accuracy.
Real-World Applications
Drug Discovery: AlphaFold assists in virtual screening, binding site prediction, and rational drug design. ESMFold accelerates candidate filtering.
Metagenomics: ESMFold rapidly predicts structures from uncultured microbial proteins, aiding in novel enzyme discovery.
Synthetic Biology: Enables de novo protein design and protein engineering by providing high-confidence structural blueprints.
Disease Research: Misfolded proteins implicated in neurodegenerative diseases can be structurally studied and targeted more effectively.
Agricultural Genomics: Structure-guided insights into plant and animal proteins improve resistance, productivity, and sustainability.
The emergence of AlphaFold and ESMFold marks a new era in molecular biology, where AI takes the reins in unraveling life’s deepest secrets. AlphaFold sets the gold standard for precision in protein modeling, while ESMFold democratizes structure prediction through speed and accessibility.
Looking ahead, innovations like AlphaFold-Multimer (for protein complexes) and ESM3 (next-gen language models) promise even deeper insights. Together, AlphaFold and ESMFold don’t just solve structures—they reshape the way we understand biology, medicine, and the molecular blueprint of life.
Subscribe To Our Newsletter
All © Copyright reserved by Accessible-Learning Hub
| Terms & Conditions
Knowledge is power. Learn with Us. 📚


