
AlphaFold 3 vs. ESMFold: Which AI Model Leads in Protein Structure Prediction?
A detailed comparison of AlphaFold 3 vs. ESMFold, two groundbreaking AI models for protein folding. Learn their differences, strengths, use cases, and impact on drug discovery, genomics, and biotechnology.
COMPANY/INDUSTRYAI/FUTUREHEALTH/DISEASE
Sachin K Chaurasiya
3/18/20254 min read

Protein folding is a cornerstone of biology, playing a crucial role in understanding diseases, designing drugs, and engineering new biomolecules. Two major AI-driven models—AlphaFold 3 and ESMFold—have revolutionized this field. But how do they compare? This article explores their strengths, differences, and impact on scientific research.
What Are AlphaFold 3 and ESMFold?
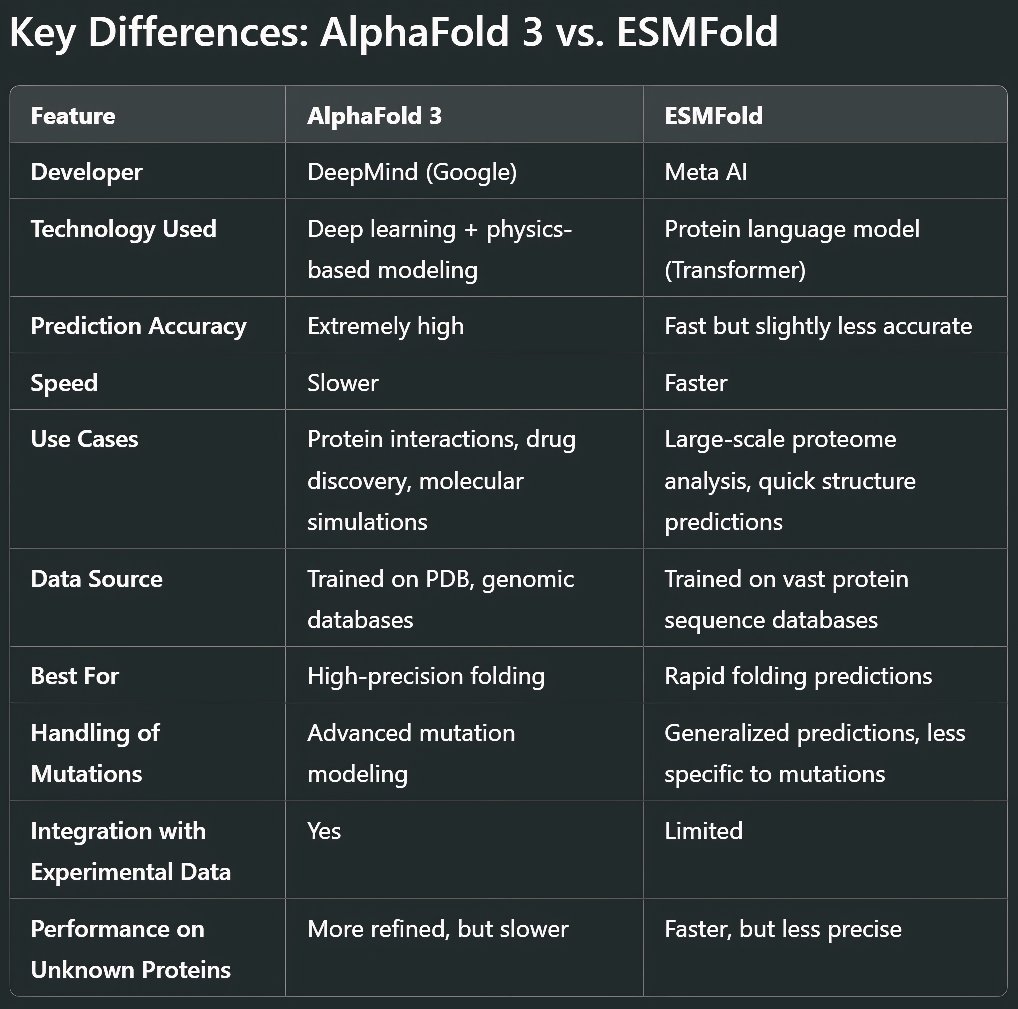
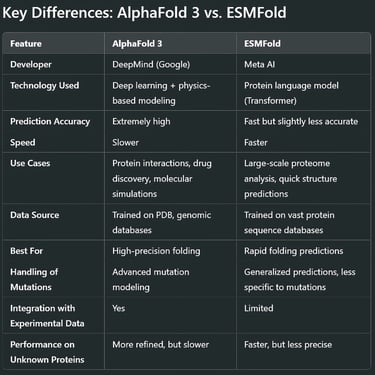
AlphaFold 3 (By DeepMind)
Developed by DeepMind, AlphaFold 3 is the latest iteration of the groundbreaking AlphaFold series. Building on its predecessors, it refines structure prediction for proteins, protein-ligand complexes, and even RNA interactions. It integrates physics-based modeling with deep learning for improved accuracy.
Enhanced modeling of protein-ligand interactions, crucial for drug discovery.
Improved understanding of RNA and DNA interactions, aiding genetic research.
Better handling of post-translational modifications, such as phosphorylation and glycosylation, which are critical for cellular functions.
Integration of cryo-EM and experimental data, leading to improved structural resolutions.
Improved computational efficiency, reducing time and resource costs for complex predictions.
ESMFold (By Meta AI)
Meta AI's ESMFold takes a different approach. Instead of AlphaFold’s structure-first training, ESMFold is based on protein language models (like GPT for proteins). It uses a large-scale transformer network to predict structures from amino acid sequences faster than AlphaFold, but sometimes at the cost of precision.
Superior scalability, allowing the analysis of entire proteomes within hours.
Rapid screening for functional proteins, which accelerates enzyme engineering and synthetic biology.
Integration with large-scale genomic studies, making it useful for evolutionary biology.
Ability to predict structures for previously unknown proteins, expanding knowledge in biodiversity and synthetic biology.
Improved fine-tuning capabilities, allowing researchers to adapt models for specific datasets and organisms.
How Do They Work?
AlphaFold 3: AI + Physics-Based Modeling
Amino Acid Input → Feeds protein sequences into a deep learning model.
Attention Mechanisms → Learns residue-residue interactions using evolutionary data.
Physics-Based Refinements → Uses molecular dynamics simulations for enhanced accuracy.
Cryo-EM and Experimental Data Integration → Further improves resolution for real-world applications.
Final Prediction → Outputs atomic-level structures with high confidence.
ESMFold: Language Model for Proteins
Amino Acid Input → Converts sequences into tokenized embeddings (like a language model).
Transformer Network → Predicts folding patterns based on contextual protein knowledge.
Structure Output → Generates a 3D model rapidly, but with variable precision.
Scalability Optimization → Allows high-throughput processing of thousands of proteins.
Strengths and Limitations
AlphaFold 3: Strengths
✔ Unmatched Accuracy: Predicts highly precise structures, even for complex biomolecules.
✔ Protein-Protein Interactions: Can model how proteins interact, aiding in drug discovery.
✔ Experimental Validation: Widely trusted by biologists for research applications.
✔ Handles Complex Mutations: Useful for predicting how genetic mutations affect protein function.
✔ Integration with Cryo-EM: Increases real-world applicability in structural biology.
AlphaFold 3: Limitations
✖ Slower Computation: Takes longer to generate predictions.
✖ Limited Ligand Predictions: Still evolving in modeling protein-small molecule interactions.
✖ Requires High Computational Power: Needs advanced GPUs and cloud computing resources.
ESMFold: Strengths
✔ Super-Fast Predictions: Ideal for large-scale studies, screening thousands of proteins.
✔ Scalability: Useful for mapping proteomes across different species.
✔ Lower Computational Cost: Requires less hardware compared to AlphaFold.
✔ Better for High-Throughput Analysis: Ideal for AI-driven bioinformatics pipelines.
✔ Effective for Unknown Protein Families: Expands the ability to predict structures for non-cataloged proteins.
ESMFold: Limitations
✖ Lower Precision: Can struggle with complex protein folding scenarios.
✖ Lacks Structural Refinement: Doesn’t integrate physics-based corrections.
✖ Not as Effective for Drug Discovery: Less suitable for modeling molecular docking and ligand binding.
FAQ's
What is the key difference between AlphaFold 3 and ESMFold?
AlphaFold 3, developed by DeepMind, uses deep learning with physics-based modeling for highly accurate protein structure predictions. ESMFold, by Meta AI, leverages protein language models (transformers) for faster but slightly less precise predictions.
Which AI model is better for drug discovery?
AlphaFold 3 is better for drug discovery due to its ability to model protein-ligand interactions, post-translational modifications, and high-resolution protein folding. ESMFold, while fast, lacks the same level of structural refinement.
How does ESMFold achieve faster predictions than AlphaFold 3?
ESMFold utilizes a transformer-based language model similar to GPT, which allows it to predict protein structures without relying on evolutionary data. This speeds up predictions, making it suitable for large-scale proteome analysis.
Can AlphaFold 3 and ESMFold predict unknown protein structures?
Yes, both models can predict unknown protein structures, but ESMFold is better suited for large-scale, rapid predictions, whereas AlphaFold 3 provides higher accuracy when dealing with novel or complex structures.
Which AI model requires more computational resources?
AlphaFold 3 requires higher computational power due to its physics-based refinements and complex modeling. ESMFold, on the other hand, is lighter and faster, making it more accessible for large-scale studies.
Are these models open-source and freely available?
Yes, both AlphaFold 3 and ESMFold are open-source. Researchers can access them to study protein folding, drug design, and genomics. However, AlphaFold 3 requires powerful hardware for optimal performance.
How accurate is AlphaFold 3 compared to experimental methods?
AlphaFold 3 approaches experimental-level accuracy for many proteins, but it is still not a replacement for lab-based techniques like X-ray crystallography or cryo-EM. It is, however, a valuable tool for accelerating research.
What are the main applications of AlphaFold 3 and ESMFold?
AlphaFold 3: Drug discovery, protein-protein interactions, molecular simulations, genetic research.
ESMFold: Large-scale proteome mapping, high-throughput protein analysis, synthetic biology.
Can these models help in understanding genetic diseases?
Yes! Both AlphaFold 3 and ESMFold can predict how genetic mutations affect protein structure, aiding in understanding diseases like cancer, Alzheimer’s, and genetic disorders.
What is the future of AI-driven protein folding?
The future involves hybrid AI models, quantum computing integrations, real-time protein simulations, and personalized medicine applications. AI-driven biology is set to revolutionize healthcare, drug development, and synthetic biology.
Future of AI in Protein Folding
Both AlphaFold 3 and ESMFold represent a paradigm shift in computational biology. The next wave of AI-driven models may combine deep learning, quantum mechanics, and real-time lab data, pushing the boundaries of molecular biology even further.
Future trends include:
AI-powered enzyme engineering for green energy solutions.
Real-time protein folding simulations to study diseases in action.
Integration with CRISPR and gene editing for personalized medicine.
Hybrid AI models combine AlphaFold and ESMFold approaches for even greater speed and accuracy.
AI-assisted drug discovery pipelines, leading to faster and more cost-effective therapeutics.
Subscribe To Our Newsletter
All © Copyright reserved by Accessible-Learning Hub
| Terms & Conditions
Knowledge is power. Learn with Us. 📚


